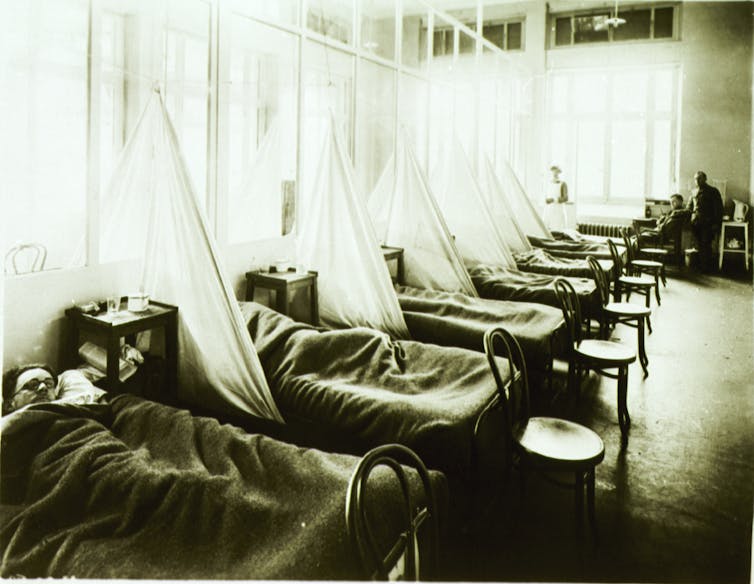
In the early 20th century, the leading cause of death was infectious disease. Epidemics erupted with little warning, seemingly out of the blue. When the “Great Influenza” struck in 1918, it killed thousands of people a week in American cities and spread like wildfire around the globe. My great aunt, still a teenager, and living in the San Francisco area, was one of its estimated 50 to 100 million victims worldwide.
Neither public health authorities nor medical researchers understood that it was a virus that caused the 1918 pandemic – most of the world at that time didn’t even know what a virus was. A century later, death due to infection is much less common, thanks to public health efforts and improved medical technology and expertise. Once common diseases are now rare. Nonetheless, infectious disease specialists like me still fear the emergence of viral diseases that we will not be able to control, including influenza.
My laboratory, along with others around the world, is working to understand how and why new influenza viruses may grip us again. To do so, we need to go far beyond human hospitals and into the wild, where viruses persist in animal populations. As disease ecologists, we aim to understand the dynamics of pathogens in the environment and their interactions with hosts. By understanding more about what’s happening with viruses in animals, we believe we can be better prepared to evaluate, predict and respond if an infection spills over to humans, making people sick.
Identifying the invisible, infectious virus
Until well into the 1930s, the “Spanish flu” was mistakenly thought to be a bacterial infection, with Haemophilus influenzae commonly blamed. This bacterium is a pathogen in its own right and may have contributed heavily to the 1918 pandemic’s death toll – but it was a secondary infection in many of the severe cases, not the original cause of victims’ illnesses.
Researchers had only identified viral particles for the first time less than 30 years before the height of the flu pandemic and the fledgling field of virology was just beginning to identify them as causes of disease in plants and animals. Scientists were only first able to visualize a virus, the tobacco mosaic virus, after the 1931 invention of the electron microscope. Though the technology, knowledge and pace of research was different early in the 20th century, why did the discovery of influenza virus take so long?
The answer, it seems, lay at least in part in people’s naiveté about the relationship between animals, the environment and human disease. In 1918, veterinarian J.S. Koen noted a very similar disease to influenza in pigs. Yet, it wasn’t until 1931 that researcher Richard Shope identified a filterable agent, smaller than bacteria, as the cause of the disease in pigs and demonstrated the transmission of an influenza virus. That work spurred the description of human influenza virus in 1933.
The tools of molecular biology, including nucleic acid sequencing, developed through the latter half of the 20th century, finally helped open the vault on the origins of the 1918 pandemic. In 2005, through a combination of sleuthing and sequencing of the viral genome, Jeffrey Taubenberger and a team of researchers pieced together the genetic sequence of the deadly 1918 virus, using viruses collected from the corpses of soldiers and other bodies preserved in the Arctic permafrost who died during the pandemic.
They were able to connect the origins and evolution of the 1918 pandemic with viruses that circulate in other animals, particularly those from birds and the pigs examined by Dr. Koen. Just as seen in more recent outbreaks of new influenza viruses, the 1918 pandemic traced its origins to virus strains circulating in nature.

Natural world a reservoir for human disease
The critical insight that led to the work reconstructing the 1918 virus had come in the 1970s. Led by the determination of virologist Rob Webster, researchers realized that influenza viruses are rampant in the natural world, particularly in waterfowl. In birds and possibly other animals, influenza viruses are able to replicate and transmit to new hosts without causing any severe disease. On rare occasions, given the right circumstance, this new host is a different species. This cycle, common in many pathogens, is an important part of how virus is maintained in nature and explains how animals can be a reservoir for novel influenza viruses that can cause human illnesses.
As researchers have sequenced the influenza viruses found in ducks and other birds, as well as people, swine and other animals, a picture of viral ecology based in nature has come into focus. Birds serve as a reservoir for a vast diversity of influenza viruses to which all the major human pandemics trace their origin. People were largely unaware that at the same time as the 1918 flu pandemic, pigs were sick with the disease and influenza viruses were also causing ongoing fowl plague epidemics. Exactly how and where the 1918 virus entered the human population remains controversial. But the realization that influenza virus happily exists in a wild animal reservoir has influenced the way scientists study flu – and moreover, emerging disease of any sort.
This understanding is also part of what underlies the One Health movement – the concept that the health of humans is entwined with the health of animals and of the environment. The One Health and Evolutionary Medicine initiatives are forging collaborations between medical doctors, veterinarians, ecologists, environmental researchers and those in many other fields to describe the connections among environmental change, animals and human health.

Watching the wild world to protect human health
We now know that a full 60 percent of human infectious diseases are spread from animals. In the past 20 years, that awareness has resulted in stronger efforts at influenza surveillance worldwide and the identification of several other influenza viruses that threaten public health. In my lab’s work, we endeavor to describe the ecology and natural history of influenza virus in animals to understand how new viruses arise and what the risk is of spillover into new hosts where they may cause disease.
For instance, human activity – such as the existence of open landfills, habitat destruction or farming practices – can attract or force animals to crowd into spaces they normally may not. When interactions between species and the environment are disrupted in this way, how does it affect the circulation, evolution and movement of influenza viruses or other pathogens that those animals host? Changes in the ecology of pathogens in the wild are what most frequently leads to spillover into human populations and disease outbreak.

Following an epidemic of seal deaths in 2011 in New England, our broad group of collaborators has spent cold winter days sampling seals, where we’ve discovered evidence of persistent circulating influenza viruses. These results are leading us to explore how influenza is affecting the seals, but also what the impact of a rapidly expanding seal population will be on the virus. If seals are a mammalian reservoir more commonly infected than we knew, their populations may affect influenza disease ecology.
Surveillance and research work like that on influenza and its animal hosts has led to more aggressive efforts to stamp out emerging infections before they become human pandemics. It gives biomedical researchers a head start on characterizing possible pandemic viruses to understand their potential impact. And public health workers gain new insights on prevention and control of infection.
That information may be crucial in identifying and containing the next pandemic virus. The One Health community’s experience with influenza has informed how scientists try to understand and prevent the spread of other diseases, including SARS, Ebola and Zika. Researchers were quick to chase after the animal source of SARS and are still hard at work to identify reservoir hosts and understand the disease ecology of the Zika and Ebola viruses.
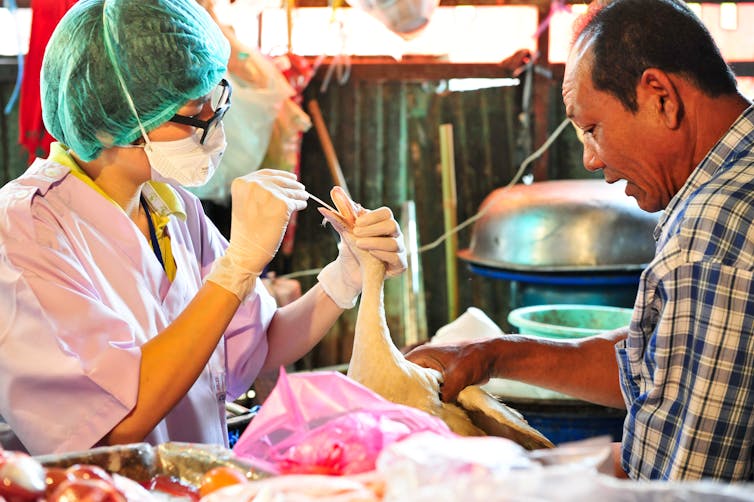
Over one hundred years after the “Great Influenza,” there’s still much to learn to lessen the risk of a repeat of 1918. In the last 10 years, thanks to the efforts of many researchers worldwide, including a renewed effort funded by the U.S. National Institutes of Health, the pace of sequencing influenza viruses has leapt forward. Scientists are beginning to understand the true diversity of influenza virus, not only in birds, but in other animals as well.
Efforts at producing a universal vaccine to prevent influenza infection in humans show promise. But the ability to test those vaccines and to prepare for and predict emerging strains will not be complete without a strong understanding of the origin, movement and risk of viruses circulating in the animals and environment around us. With better understanding of these ecological connections coming from continued research, we hope we can be better prepared for the next pandemic.

