Wastewater surveillance hit the big time during the height of the COVID-19 pandemic, when officials started using this technique to monitor local virus levels. But my colleagues and I had been exploring wastewater’s promise as a public health tool years before anyone had heard of SARS-CoV-2.
My environmental virology lab based at Michigan State University has been in a partnership with the city of Detroit and the Great Lakes Water Authority since 2017, when we started testing municipal wastewater from Wayne, Oakland and Macomb counties to survey viral diseases in the Greater Detroit community.
Imagine you want to identify a potential emerging infectious disease in an urban area before it becomes an outbreak. Could you collect clinical samples from everyone in the community on a regular basis and test them all for every possible virus? No, that’s an impossible task.
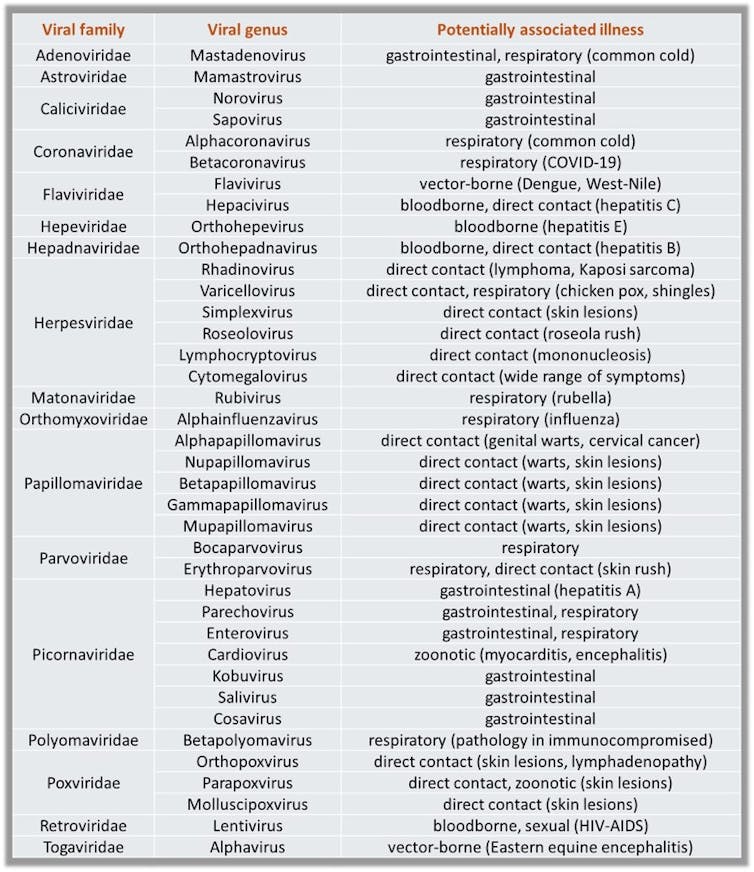
Instead, we collected municipal wastewater, as a representative composite sample from the community, and tested that with advanced molecular methods to reveal endemic and emerging virus-related diseases circulating in the area. We identified viral genomes related to multiple gastrointestinal, respiratory, blood-borne and vector-borne diseases excreted by the population. We identified herpesviruses, including rare species, and we identified hepatitis A outbreak peaks via wastewater analysis that appeared before peaks in clinical samples.
The ongoing project in Detroit shifted to monitor SARS-CoV-2 in 2020. Our team, which then expanded to include the local health departments, practicing engineers and the Michigan Department of Health and Human Services, was able to identify COVID-19 peaks in the metro Detroit population five weeks before the delta variant surge was visible in positive COVID test data.
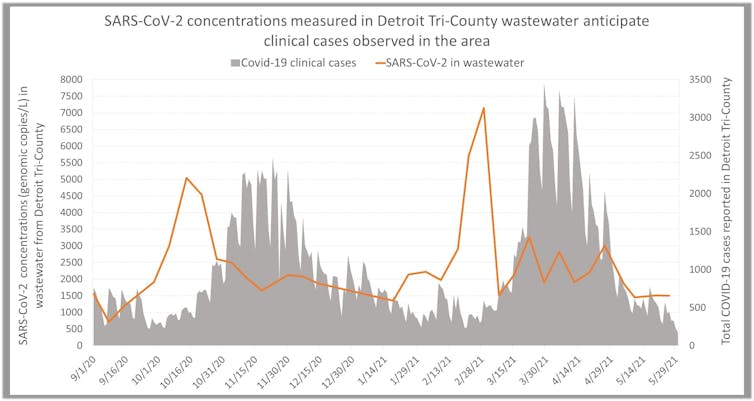
Our team is continuing to expand and improve our methodology to screen for the presence of endemic and emerging communicable diseases circulating in the metro Detroit population and to predict when and where surges will occur.
Data hidden in what goes down the drain
Wastewater is a mixture that contains anything that goes down the drain from the toilet, the shower, the laundry, the dishwasher or the sink. In most cities, stormwater and water used in industrial processes also end up in the wastewater.
Municipal wastewater contains viruses and other pathogens excreted by anyone with an infection, even at the early stages before symptoms develop.
These microorganisms are diluted in a large amount of water and mixed up with soap, personal care products, road runoff and many other chemicals and impurities. The drop of blood or saliva that a clinic would test for infectious agents is much more concentrated and much less contaminated with other molecules.

Looking for emerging human viruses in wastewater is like trying to find a needle in a haystack. We concentrate and isolate viruses in the wastewater samples. We then extract any DNA or RNA that’s in the samples and analyze it with molecular methods such as PCR and next-generation sequencing, looking for genes related to viruses. Those genetic codes indicate which viral infections are present in the population that produced the wastewater.
Beyond confirming a pathogen’s presence
It’s one thing to identify that a certain virus is present in a community. But it’s a trickier task to figure out how a particular wastewater level translates to how many people are sick or to compare infection levels between communities. How quickly will a surge of people start turning up sick once a wastewater peak is spotted?
To predict the timing of outbreak peaks, scientists consider multiple variables, including how much virus a typical patient sheds, how long people shed the pathogen after infection, the onset and duration of clinical symptoms, and how long the pathogen has been in the wastewater collection pipes. Using all these factors, we relate wastewater readings to metrics of clinical disease.
In addition to complex models that predict variation over time and peaks of clinical cases, we developed simple methods for data analysis that can be used for decision-making by public health officials. We also tracked the time lag between SARS-CoV-2 concentrations in wastewater and clinical metrics.
An important step is connecting a particular viral reading in wastewater with the number of people who live in the area served by that sewer system. We use molecules produced by the human body to calculate how many people’s wastewater feeds into our sample. For example, using metabolites such as creatine, 5-HIAA and xanthine, we calculated per capita COVID-19 trends by relating their levels with viral levels. This process allows for comparisons between communities.
Screening for the next outbreak
Ultimately, public health officials want to get ahead of any looming outbreak. To this end, our team developed a ranking system for prioritizing which reportable diseases future wastewater surveillance should focus on.
Clinicians are legally required to report dozens of diseases – ranging from chickenpox and COVID-19 to meningitis and measles – to the health department when they’re diagnosed. Keeping track of these reportable diseases lets officials count and record cases and trends.
Our ranking system is one of the first of its kind. We use 12 factors, including clinical trends in specific geographic locations and knowledge of how contagious particular germs are. Knowing which communicable diseases are heating up in a particular community allows officials to prioritize resources and efforts toward monitoring and preventing their spread.
In the Detroit area, for instance, we found that certain diseases, including some sexually transmitted infections, ranked higher than in the state of Michigan as a whole. It makes sense to allocate resources to addressing them in a targeted way.
We also developed a sequencing and bioinformatics protocol, a step-by-step process that screens for potential viral-related sequences that may be circulating in the community. It’s a way to broaden surveillance beyond the usual diseases that medical providers must report to officials when diagnosed. This tool can provide an early warning to officials if something new or unexpected shows up in wastewater.
Solidifying the science of wastewater surveillance
Wastewater surveillance has proved itself as an epidemiological tool. But even though wastewater-based epidemiology has made significant advances in technology, methods and applications, more needs to be done to integrate the multiple efforts across the nation. For accurate outbreak forecasting, wastewater surveillance databases should be integrated with clinical data metrics, behavioral, social and demographic information, as well as data that indicates population mobility. Close partnerships with local health departments are crucial, since local epidemiologists are the ones who will use surveillance efforts to make decisions.

